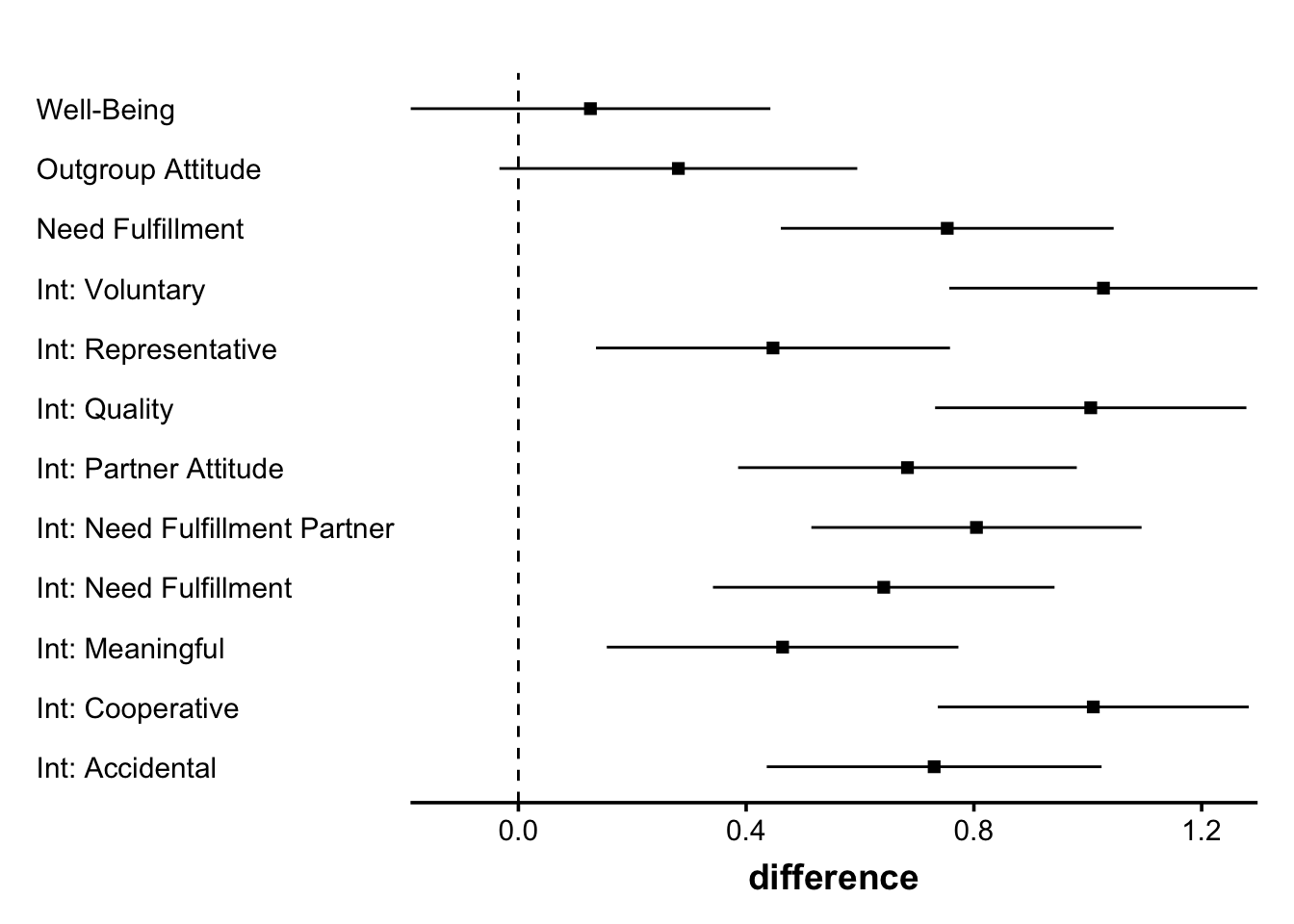
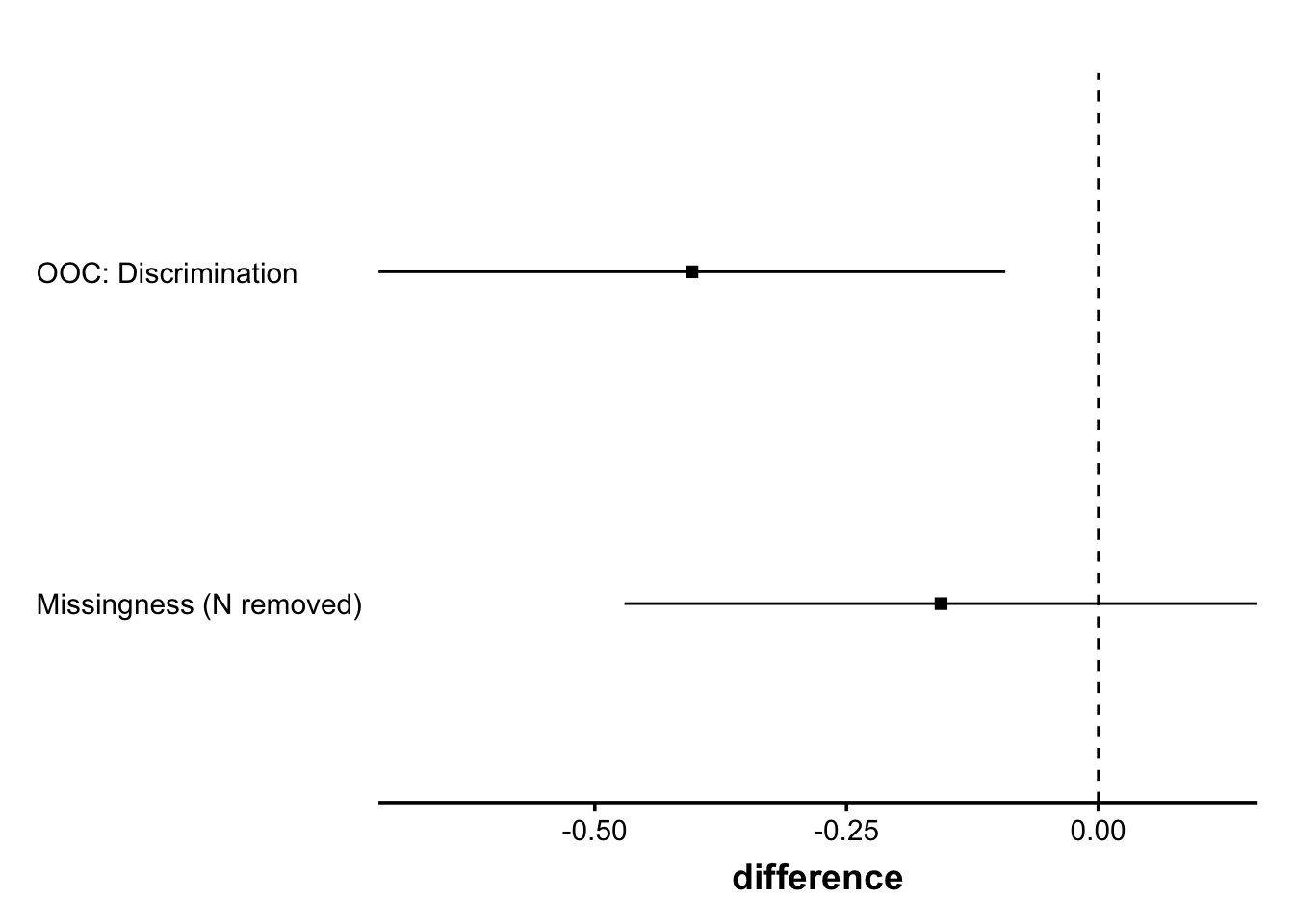
Bringmann, L. F., & Eronen, M. I. (2018). Don’t blame the model:
Reconsidering the network approach to psychopathology.
Psychological Review,
125(4), 606–615.
https://doi.org/10.1037/rev0000108
Eronen, M. I., & Bringmann, L. F. (2021).
The Theory Crisis in Psychology: How to Move Forward. 779–788.
https://doi.org/10.1177/1745691620970586
Kaufman, L., & Rousseeuw, P. J. (Eds.). (1990).
Finding Groups in Data. John Wiley & Sons, Inc.
https://doi.org/10.1002/9780470316801
Keogh, E., & Kasetty, S. (2003). On the
Need for
Time Series Data Mining Benchmarks:
A Survey and
Empirical Demonstration.
Data Mining and Knowledge Discovery,
7(4), 349–371.
https://doi.org/10.1023/A:1024988512476
Kittler, J., Hatef, M., Duin, R. P. W., & Matas, J. (1998). On combining classifiers.
IEEE Transactions on Pattern Analysis and Machine Intelligence,
20(3), 226–239.
https://doi.org/10.1109/34.667881
Kogan, J., Nicholas, C. K., & Teboulle, M. (Eds.). (2006). Grouping multidimensional data: Recent advances in clustering. Springer.
Monden, R., Rosmalen, J. G. M., Wardenaar, K. J., & Creed, F. (2022). Predictors of new onsets of irritable bowel syndrome, chronic fatigue syndrome and fibromyalgia: The lifelines study.
Psychological Medicine,
52(1), 112–120.
https://doi.org/10.1017/S0033291720001774
Niennattrakul, V., & Ratanamahatana, C. A. (2007). Inaccuracies of
Shape Averaging Method Using Dynamic Time Warping for
Time Series Data. In D. Hutchison, T. Kanade, J. Kittler, J. M. Kleinberg, F. Mattern, J. C. Mitchell, M. Naor, O. Nierstrasz, C. P. Rangan, B. Steffen, M. Sudan, D. Terzopoulos, D. Tygar, M. Y. Vardi, G. Weikum, Y. Shi, G. D. van Albada, J. Dongarra, & P. M. A. Sloot (Eds.),
Computational Science – ICCS 2007 (Vol. 4487, pp. 513–520). Springer Berlin Heidelberg.
https://doi.org/10.1007/978-3-540-72584-8_68
Rousseeuw, P. J. (1987). Silhouettes:
A graphical aid to the interpretation and validation of cluster analysis.
Journal of Computational and Applied Mathematics,
20, 53–65.
https://doi.org/10.1016/0377-0427(87)90125-7
Syakur, M. A., Khotimah, B. K., Rochman, E. M. S., & Satoto, B. D. (2018). Integration
K-
Means Clustering Method and
Elbow Method For Identification of
The Best Customer Profile Cluster.
IOP Conference Series: Materials Science and Engineering,
336, 012017.
https://doi.org/10.1088/1757-899X/336/1/012017